The modules are customizable extensions to the OpenBioMaps web application. There are project-level modules (e.g.: postgres repository creation, photo manager) and there are also modules specific to individual data tables (e.g.: text filters for map page, CSV export).
The use of modules can be assigned to different users or user groups.
The modules are linked to module hooks in the application, which are mostly located on the map page and profile tab. Most modules can be configured with simple parameters (JSON), but some modules have a custom administrative interface.
Module administration¶
The modules can be enabled and configured on the project_administration -> modules page.
Add a custom module¶
You can upload your modules and add them to your project. To develop a module, check out the example modules in modules/examples/.
Module access¶
You can add each module to your list several times. This allows us to give each module multiple access levels. This is important for modules where we want to give different access to different users or groups, for example, the allowed_columns module. Another example is that if we have multiple data tables, we can specify for each table separately which column values we can filter based on when querying, for example, the text_filter module. In the Access column, we can choose whether our settings are public (everybody) or only logged-in users (logged users) can use the given option. In the Group Access column, we can further refine our access options by selecting our predefined groups or even assigning specific persons to a particular setting.
Drop a module¶
Currently, there is no such option.
Turn off a module¶
Once we have our list of modules, we can switch each module on and off.
Parameters for modules¶
Most modules can be directly parameterized with JSON format parameters. Some modules have their administrative tab, through which administrative tasks related to the module can be performed. An example is the box_load_selection module.
Project level modules¶
box_load_selection¶
Allows you to upload your spatial shapes (points, lines, polygons). These are usually SHP files, but can also be other standard spatial data formats.
The uploaded spatial shapes can be used by users for data queries or data uploads. In both cases, the spatial object can be used to refer to the name of the shape, either to spatially delimit the data query or to specify the spatial location of the uploaded data record.
Uploaded spatial objects can be shared with other users, who can decide whether they want to use these shapes. By default, newly uploaded spatial shapes are not visible to other users. To use objects uploaded by others, you need to allow queries or data uploads. Project owners can set these permissions for each user for each spatial object.
Users can access spatial shapes via the “Shared Geometries “ module block found in the module section at the bottom of their profile page. Project administrators can make these settings in their administration interface of the box_load_selection module in the module settings tab.
Once the module is activated, the “Spatial query “ box appears on the Map page. Here you will see a drop-down list of the names of the spatial shapes available to you, from which you can perform a spatial query on the database. In the case of a polygon, you can choose to query only the data that are inside the polygon or also the data that fall under the edges of the polygon.
For web and file uploads, if the “obm_geometry “ column type is used for coordinate capture, clicking on the map marker icon in a pop-up window will display a drop-down menu with “geometry from list “, in which you can select the required spatial shape by name, for which the application will load the WKT coordinate into the corresponding geometry field of the upload form.
The spatial shapes available for upload will also be downloaded by the mobile app and will be semi-transparently drawn on the map in the upload forms, labelled with their names.
No parameters
photos¶
Photo or other attachment boxes.
No parameters
create_pg_user¶
After enabling the module, the Create Postgres user box will appear on the profile page.
By enabling the module, users who are granted the right to use the module will be able to create their own Postgres user.
By default, the module creates a POSTGRES user with limited access, who can read all database tables in your project. He can only connect to the database from one client program at a time, and his access automatically expires after one year.
The created Postgres user will be added to the *project_name*_user group, which is the Postgres Group automatically created by the system. With Postgres admin access, you can set additional rights for certain users, e.g. write access to certain tables.
Users can renew their access at any time.
The Postgres user can be used to connect to the database from QGIS, for example. An example of how to set this up:
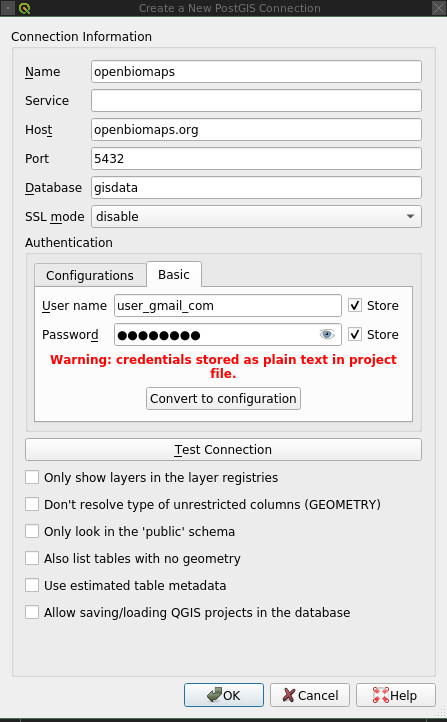
No parameters
computation¶
No parameters
custom_filetype¶
Custom file preparation. E.g. observado style CSV
No parameters
taxon_meta¶
No parameters
Table level modules¶
additional_columns¶
If a database consists of several data tables, they can be linked by different variables.
When queried, all data for an identifier is queried. This function can be ignored by checking “ignore table JOINS “ on the map page.
For example, in some burrow projects we keep the data for parents and offspring in separate tables, if we want to get all the data for a burrow, we can specify the “odu_asonosito “ column as the “join” variable.
use it together with the join_tables module
return with an array: co [0] columns array c [1] column name assoc array Parameters: ["column names"]
allowed_columns¶
Here you can set which column should be visible at different access levels.
It can be used if the data table has a “rules “ table and the basic access level of the project is not public.
- Parameters:
{
"for_sensitive_data": [...] The columns we want to make visible. Does not show the geometry associated with the data. "for_no-geom_data": [...]
Columns that you want to make visible. "for_general": [...] For the columns, we want to make them visible. }
bold_yellow¶
Column names in yellow bold in the result lists. After a query, column names in bold yellow appear in the detailed description attached to the “Drop-down list “ table.
This module is also used to specify which data should be displayed in the “Recorded data “ summary labels in the mobile application.
- Parameters:
[“column names”]
box_load_coord¶
On the map page, a “position “ block will appear below the map. If you move the cursor around the map, you will see that the coordinate in the “position “ block is constantly changing, tracking the position of your cursor on the map.
Also in the “position “ block, if you type in the latitude and longitude, clicking on the little black “lollipop “ will display your point on the map.
Parameters:
{
"wgs84": "4326",
"eov": "23700"
}
box_load_last_data¶
Create Quick queries option on the map page on the right side of the map. There are three options to choose from:
last own upload,
last upload (anyone’s)
last uploaded rows.
On the module page you can set the amount of last uploaded rows to be queried. For the other two options, the module always returns 1 row.
- Parameters:
[“Number of records in last uploads, default is 10”]
box_custom¶
Custom box on the map page - only user-defined version exists. The custom module has to be placed in the local/includes/modules/ folder.
Parameters:
[a file's basename includes/modules/private folder. E.g. hrsz_query]
Where hrsz_query_Class is a class in local/includes/modules/hrsz_query.php file.
This Class should include at least print_box() and print_js() functions.
identify_point¶
Identify one or more points on the map.
Display in a small bubble some information about the data point that has been previously set.
Parameters: [column names]
custom_notify¶
Creates custom Postgres-based notify events. This is just an idea, the module is not ready.
No parameters
custom_data_check¶
Custom data checks of upload data.
No parameters
download_restricted¶
Admin-controlled download authorization
No parameters
extra_params¶
Extra input parameters for forms.
grid_view¶
View data on a custom polygon grid. E.g UTM 2.5km, UTM 10KM, KEF grid, snap to grid, …
When using grid modules, the original geometry of the data is also taken from the grid module.
Methods: print_box(), default_grid_geom(), get_grid_layer()
Parameters: layer_options
Parameters example:
{
"layer_options": [
"kef_5 (layer_data_grid)",
"original (layer_data_points)"
]
}
The layer_options contains the data view modes and their options that users can choose, i.e. which layer should be associated with which MapServer layer.
The name of the MapServer layer should be given in parentheses. The string before the parentheses is the corresponding column name of the YOURTABLE_qgrids table, i.e. the column containing the desired grid geometry is linked to the MapServer layer that manages it.
The grid layer is a polygon layer, so you need to create a polygon layer in the Mapfile. Eg: layer_data_grid
The module will create a YOURTABLE_qgrids table for you if it does not already exist, which you can modify according to the grid types you want to apply.
The module will also create update_grid_geoms trigger and set the comments for you, but most probably you need to modify them.
In the YOURTABLE_qgrids table, set the visible names of the layers in the comment field. These will be the names that users will see in the options list.
COMMENT ON COLUMN public.YOURTABLE_qgrids.original IS 'original';
COMMENT ON COLUMN public.YOURTABLE_qgrids.kef5 IS 'KEF 5x5';
Example trigger functions:
Trigger on YOURTABLE_qgrids. Arguments are in the function:
CREATE TRIGGER update_grid_geoms BEFORE INSERT OR UPDATE ON public.tytoalba_qgrids FOR EACH ROW EXECUTE PROCEDURE public.update_qgrid_geoms_arg('0.1', '0.1', 't', 't', 't', 't', '0.05');
Trigger on YOURTABLE table:
CREATE TRIGGER qgrids BEFORE INSERT OR DELETE OR UPDATE ON public.YOURTABLE FOR EACH ROW EXECUTE PROCEDURE insert_originalgeom_into_qgrids()
Function insert_originalgeom_into_qgrids()
BEGIN
IF tg_op = 'INSERT' THEN
EXECUTE format('INSERT INTO %I_qgrids (row_id,original) SELECT %L,%L::geometry',TG_TABLE_NAME,NEW.obm_id,NEW.obm_geometry);
RETURN NEW;
END IF;
IF tg_op = 'UPDATE' THEN
EXECUTE format('UPDATE %I_qgrids SET "original"=%L::geometry WHERE row_id=%L', TG_TABLE_NAME,NEW.obm_geometry,NEW.obm_id);
RETURN NEW;
END IF;
IF tg_op = 'DELETE' THEN
EXECUTE format('DELETE FROM %I_qgrids WHERE row_id=%L',TG_TABLE_NAME,OLD.obm_id);
RETURN OLD;
END IF;
END;
Function update_qgrid_geoms_arg()
DECLARE
snap_x numeric := TG_ARGV[0];
snap_y numeric := TG_ARGV[1];
kef5 boolean := TG_ARGV[2];
utm10 boolean := TG_ARGV[5];
snap boolean := TG_ARGV[6];
snap_polygon boolean := TG_ARGV[7];
snap_polygon_size numeric := TG_ARGV[8];
BEGIN
IF tg_op = 'UPDATE' THEN
IF kef5 THEN
EXECUTE FORMAT('SELECT geometry FROM shared."kef_5x5" WHERE st_within(%L::geometry,geometry)',NEW.original) INTO NEW."kef_5";
END IF;
IF snap THEN
EXECUTE FORMAT('SELECT st_SnapToGrid(%L::geometry,%L,%L)',NEW.original,snap_x,snap_y) INTO NEW."snap";
END IF;
IF snap_polygon THEN
EXECUTE FORMAT('SELECT st_expand(st_SnapToGrid(%L::geometry,%L,%L),%L)',NEW.original,snap_x,snap_y,snap_polygon_size) INTO NEW."snap_polygon";
END IF;
RETURN NEW;
END IF;
IF tg_op = 'INSERT' THEN
IF kef5 THEN
EXECUTE FORMAT('SELECT geometry FROM shared."kef_5x5" WHERE st_within(%L::geometry,geometry)',NEW.original) INTO NEW."kef_5";
END IF;
IF snap THEN
EXECUTE FORMAT('SELECT st_SnapToGrid(%L::geometry,%L,%L)',NEW.original,snap_x,snap_y) INTO NEW."snap";
END IF;
IF snap_polygon THEN
EXECUTE FORMAT('SELECT st_expand(st_SnapToGrid(%L::geometry,%L,%L),%L)',NEW.original,snap_x,snap_y,snap_polygon_size) INTO NEW."snap_polygon";
END IF;
RETURN NEW;
END IF;
END;
When you are ready to prepare the _qgrids table, you need to add the existing geometries from the target table if it is not already empty.
INSERT INTO YOURTABLE_qgrids (row_id,"original") SELECT obm_id,obm_geometry FROM YOURTABLE;
UPDATE YOURTABLE_qgrids SET "snap"=foo.obm_geometry FROM (SELECT st_SnapToGrid(obm_geometry,0.13,0.09) as obm_geometry,obm_id FROM YOURTABLE) AS foo WHERE row_id=obm_id
UPDATE YOURTABLE_qgrids SET "kef_5"=foo.obm_geometry
FROM (
SELECT d.obm_id,k.obm_geometry FROM YOURTABLE d LEFT JOIN shared."kef_5x5" k ON (st_within(d.obm_geometry,k.obm_geometry) )
) as foo
WHERE row_id=foo.obm_id
In this example, the “shared”. “kef_5x5” table contains the polygons we want to use, and we also created a custom polygon called “snap” on the fly.
job_manager (validation)¶
General description:
* The job_manager (validation) module is used for managing the background processes of the project. Its parameters are the names of the jobs.
* On the admin page you can set the time of running (simplified cron style: minute hour day), and the job parameters as JSON
* Adding a new parameter will register the job in the jobs database table, and create the necessary template files in the modules/validation_modules, jobs folders.
Parameters:
* The names of the background processes
Published jobs:
observation_lists
description:
This job processes and copies the observation lists collected and uploaded by the mobile app. The observations land in a temporary table, where this job completes the obm_obsevation_list_id column, the columns of start, end, and duration of the list. If the uploaded list is not complete the list is skipped.
parameters:
* list_start_column (string): column name of list start
* list_end_column (string): column name of list end
* list_duration_column (string): column name of list duration
* only_time (boolean): store whole timestamp or just time
* time_as_int (boolean): convert time to minutes
{
"tablename": {
"list_start_column": "time_of_start",
"list_end_column": "time_of_end",
"list_duration_column": "duration",
"only_time": true,
"time_as_int": true
}
}
}
incomplete_observation_lists
description:
If the uploaded list is incomplete, this module processes it. If the difference is smaller than the tolerance value, then the list will be uploaded by the next observation_list process, but a system message is sent. In another case, when the difference is larger than the tolerance only a system message is sent, the rest has to be processed manually.
Parameters:
* mail_to (int): role_id - who will get the messages
* diff_tolerance (int): tolerance of difference
* days_offset (int): number of days to wait until the list is processed
{
"tablename": {
"mail_to": 1265,
"diff_tolerance": 2,
"days_offset": 2
}
}
join_tables¶
This module makes it possible to display joined data on the data-sheet-page. At the moment it supports only simple LEFT JOINS on one equation.
- Parameters:
- [
- {
“table”: “teszt_events”, “join_on”: [
- {
“ref_field”: “obm_id”, “join_field”: “patient_id”
}
]
}, {
“table”: “teszt_masik”, “join_on”: [
- {
“ref_field”: “obm_id”, “join_field”: “fid”
}
]
}
]
list_manager¶
No parameters
massive_edit¶
Allows bulk editing of selected data on the map page via the file upload interface
No parameters
move_project¶
Move the project to another server. This is an experimental module.
No parameters
read_table¶
Present a SQL table or an SQL view as a rollable html table. This table is available through a unique link.
Parameters:
[{"table": "schema.table",
"label": "somesthing",
"orderby": "column"}, ... ]
results_asList¶
Create foldable slides-like results output.
No parameters
results_asGPX¶
Save results as a GPX file.
Parameters:
{"name": "column",
"description": ["column-1", "column-2", ... ]}
results_asCSV¶
Save results as a csv file.
Parameters:
{"sep": ",",
"quote":"\""}
results_asJSON¶
Save results as a JSON file.
No parameters
results_asSHP¶
Results can be saved as SHP files. Separate files are created for the different geometry types. These can be downloaded in a zip archive.
No parameters
results_asKML¶
Save results as a KML file.
Parameters:
{"name": "column",
"description": ["column-1", "column-2", ... ]}
results_asStable¶
Compact results table on the map page.
- Parameters:
[column names]
results_specieslist¶
Species list summary on the map page
No parameters
results_summary¶
A summary of results.
No parameters
results_table¶
Create a full html table of the results.
No parameters
restricted_data¶
Rule-based data restriction
No parameters
text_filter¶
Text filters on the map page and for query API. Create the WHERE part of the SQL query string.
Parameters:
[
"magyar",
"obm_taxon",
"megj::colour_rings",
"obm_datum",
"obm_uploading_date",
"obm_uploader_user",
"d.szamossag:nested(d.egyedszam):autocomplete",
"d.egyedszam:values():",
"obm_files_id",
"faj::autocomplete"
]
text_filter2¶
Advanced taxon and other text filters. Create the WHERE part of the SQL query string.
Parameters:
{...}
transform_data¶
Transform fields to better reading on web tables and exports. E.g. In the result list, it can transform geometry to WKT.
Parameters:
{
"obm_geometry":"geom",
"obm_uploading_id":"uplid",
"tema":"mmm"
}
